APPLICATIONS
1.
Matrix computation for Google’s
PageRank algorithm
How to solve the matrix equation and obtain
Google solution for the textbook example
(Elements of Statistical
Learning, pp. 576-578)

>
x=c(0,1,1,0,0,0,1,0,1,0,0,0,0,0,1,0)
> L = matrix(x,4,4) #
Define a 4x4 matrix L showing linkage among web pages
> L # R
reads a matrix column by column
[,1] [,2] [,3] [,4]
[1,] 0
0 1 0
[2,] 1
0 0 0
[3,] 1
1 0 1
[4,] 0
0 0 0
> e = matrix( rep(1,4), 4, 1 ) #
Vector of 1s
> e
[,1]
[1,] 1
[2,] 1
[3,] 1
[4,] 1
> C = diag( colSums(L) ) # Diagonal matrix C showing the number
> C # of outgoing links from each web page
[,1] [,2] [,3] [,4]
[1,] 2
0 0 0 # Matrix multiplication is %*%
[2,] 0
1 0 0 # Inverse matrix is solve(C)
[3,] 0
0 1 0 # Transpose matrix is t(e)
[4,] 0
0 0 1
> A = 0.15*(e%*%t(e))/4 + 0.85*L%*%solve(C)
> A
[,1] [,2] [,3] [,4]
[1,] 0.0375 0.0375 0.8875
0.0375
[2,] 0.4625 0.0375 0.0375
0.0375
[3,] 0.4625 0.8875 0.0375
0.8875
[4,] 0.0375 0.0375 0.0375
0.0375
> eigen(A) # Computes eigenvalues and eigenvectors of
A
$values
[1] 1.000000e+00+0.00e+00i
-4.250000e-01+4.25e-01i
[3] -4.250000e-01-4.25e-01i 5.477659e-18+0.00e+00i
$vectors
[,1] [,2]
[1,] -0.64470397+0i 7.071068e-01+0.000000e+00i
[2,] -0.33889759+0i
-3.535534e-01-3.535534e-01i
[3,] -0.68212419+0i
-3.535534e-01+3.535534e-01i
[4,] -0.06489841+0i -2.175463e-17-4.017163e-18i
[,3] [,4]
[1,] 7.071068e-01+0.000000e+00i
-9.941546e-16+0i
[2,]
-3.535534e-01+3.535534e-01i -7.071068e-01+0i
[3,]
-3.535534e-01-3.535534e-01i -1.968623e-16+0i
[4,]
-2.175463e-17+4.017163e-18i
7.071068e-01+0i
> install.packages("expm”) # This package contains matrix powers
> library(expm) # “Exponentiation
of a matrix”
> A100 = A%^%100 #
Large power of matrix A
> A100 # (a
simple but not an optimal method)
[,1] [,2] [,3] [,4]
[1,] 0.3725269 0.3725269
0.3725269 0.3725269
[2,] 0.1958239 0.1958239
0.1958239 0.1958239
[3,] 0.3941492 0.3941492
0.3941492 0.3941492
[4,] 0.0375000 0.0375000
0.0375000 0.0375000
# “Converged”!
All columns are the same
> p = A100[,1] #
Take one column of A100
> p
[1] 0.3725269 0.1958239
0.3941492 0.0375000
> p=t(t(p)) #
Now R understands it as a column
> p
[,1]
[1,] 0.3725269
[2,] 0.1958239
[3,] 0.3941492
[4,] 0.0375000
#
Verify that it is a solution of p = Ap
> A%*%p
[,1]
[1,] 0.3725269
[2,] 0.1958239
[3,] 0.3941492
[4,] 0.0375000
# p is not properly normalized yet, we need e’p = N = 4
> t(e)%*%p # but we have the sum of p[i] = 1
[,1]
[1,] 1
> p*4 # This is the solution. Ranked by importance, we have
[,1] # web pages ordered as 3, 1, 2, 1
[1,] 1.4901074
[2,] 0.7832956
[3,] 1.5765969
[4,] 0.1500000
# Another way is by
iterations (standard method of solving fixed-point equations):
> p = matrix(c(1,0,0,0),4,1) # The initial vector is kind of arbitrary
> p
[,1]
[1,] 1
[2,] 0
[3,] 0
[4,] 0
> for
(n in (1:100)){ p = A%*%p } # Iterate p[next] = A*p[current]
> 4*p # Same result
[,1]
[1,] 1.4901074
[2,] 0.7832956
[3,] 1.5765969
[4,] 0.1500000
2.
Spam detection
# The Spam data with description are in https://archive.ics.uci.edu/ml/datasets/Spambase
# We can also get its
formatted version from a package “kernlab”
> library(kernlab)
> data(spam)
> dim(spam)
[1] 4601 58
> names(spam)
[1] "make"
"address"
"all"
"num3d"
[5] "our"
"over"
"remove"
"internet"
[9] "order"
"mail"
"receive"
"will"
[13] "people" "report" "addresses" "free"
[17] "business" "email" "you" "credit"
[21] "your" "font" "num000" "money"
[25] "hp" "hpl" "george" "num650"
[29] "lab" "labs" "telnet" "num857"
[33] "data" "num415" "num85" "technology"
[37] "num1999" "parts" "pm" "direct"
[41] "cs" "meeting" "original" "project"
[45] "re" "edu" "table" "conference"
[49] "charSemicolon" "charRoundbracket" "charSquarebracket"
"charExclamation"
[53] "charDollar" "charHash" "capitalAve" "capitalLong"
[57] "capitalTotal" "type"
> table(type)
type
nonspam spam
2788
1813
# LOGISTIC REGRESSION
> logreg = glm( type ~ .,
data=spam, family = "binomial" )
> prob.spam
= fitted.values(logreg)
> class.spam
= ifelse(
prob.spam > 0.5, "spam", "nonspam" )
> table(class.spam, type)
type
class.spam
nonspam spam
nonspam 2666
194
spam 122 1619
> mean(
type == class.spam )
[1] 0.9313193 #
Correct classification rate = 93% (better be validated by CV)
# What are error rates
among spam and benign emails?
> sum(
type=="nonspam" & class.spam=="spam"
) / sum( type=="nonspam" )
[1] 0.04375897
> sum(
type=="spam" & class.spam=="nonspam" ) / sum( type=="spam" )
[1] 0.107005
# Reducing the error rate for nonspam emails, with loss function L(1,0)=1,
L(0,1)=10
> class.spam
= ifelse(
prob.spam > 10/11, "spam", "nonspam" )
> table(class.spam, type)
type
class.spam
nonspam spam
nonspam 2759
674
spam 29 1139
> mean(
type == class.spam )
[1] 0.8472071
> sum(
type=="nonspam" & class.spam=="spam"
) / sum( type=="nonspam" )
[1] 0.01040172
> sum(
type=="spam" & class.spam=="nonspam" ) / sum( type=="spam" )
[1] 0.3717595
# The overall error
rate increased, and the error rate of misclassifying spam emails as nonspam increased substantially, but the error rate for
benign emails is only 1% now.
# DECISION TREES
> tr = tree( type ~ ., data=spam )
> plot(tr); text(tr);
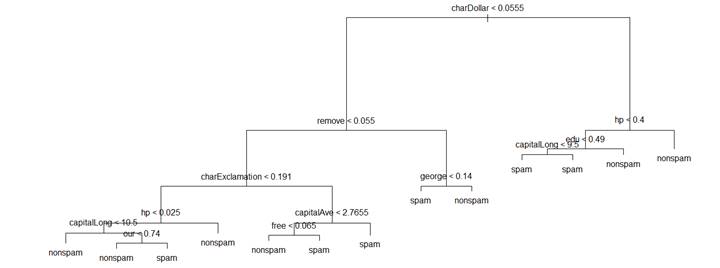
> summary(tr)
Misclassification error rate: 0.08259 = 380 / 4601
# Optimization – random forests
> library(randomForest)
> rf = randomForest( type ~ ., data=spam )
> varImpPlot(rf) # To see which variables are most
significant in a tree based classification
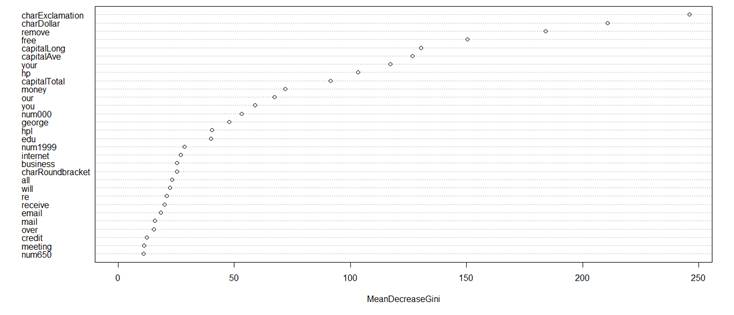
> rf
Number of trees: 500
No. of
variables tried at each split: 7 # That’s approx.
square root of the number of X-variables
OOB estimate of error rate: 4.52% # The lowest classification rate we could
get so far
Confusion matrix:
nonspam spam class.error
nonspam
2710 78 0.02797704
spam
130 1683 0.07170436
# DISCRIMINANT ANALYSIS
> library(MASS)
> LDA = lda( type
~ ., data=spam, CV=TRUE )
> table( type, LDA$class )
type nonspam spam
nonspam 2656
132
spam 394 1419
> mean( type != LDA$class )
[1] 0.114323 # This error rate
is higher
> QDA = qda( type
~ ., data=spam, CV=TRUE )
> table( type, QDA$class )
type nonspam spam
nonspam 2090
691
spam 87 1722
> mean( type != QDA$class )
[1] NA
> summary(QDA$class)
nonspam spam
NA's
2177
2413 11 # There are missing values, predictions not computed by QDA
#
We’ll remove these points
> mean( type != QDA$class,
na.rm=TRUE )
[1]
0.1694989 # No improvement with QDA; linear models
are ok
SUPPORT VECTOR MACHINES
> library(e1071)
> SVM = svm( type
~ ., data=spam, kernel="linear" )
> summary(SVM)
Number of
Support Vectors: 940
( 456 484 ) # 940 support vectors (not cleanly
classified) out of 4601 data points
> dim(spam)
[1] 4601 58
> class = predict(SVM)
> table(class,type)
type
class nonspam spam
nonspam 2663
185
spam 125 1628
> mean( class != type )
[1] 0.06737666 # Best rate so far?